About the project
FERMI: Fundamentals for a faster protein transition.
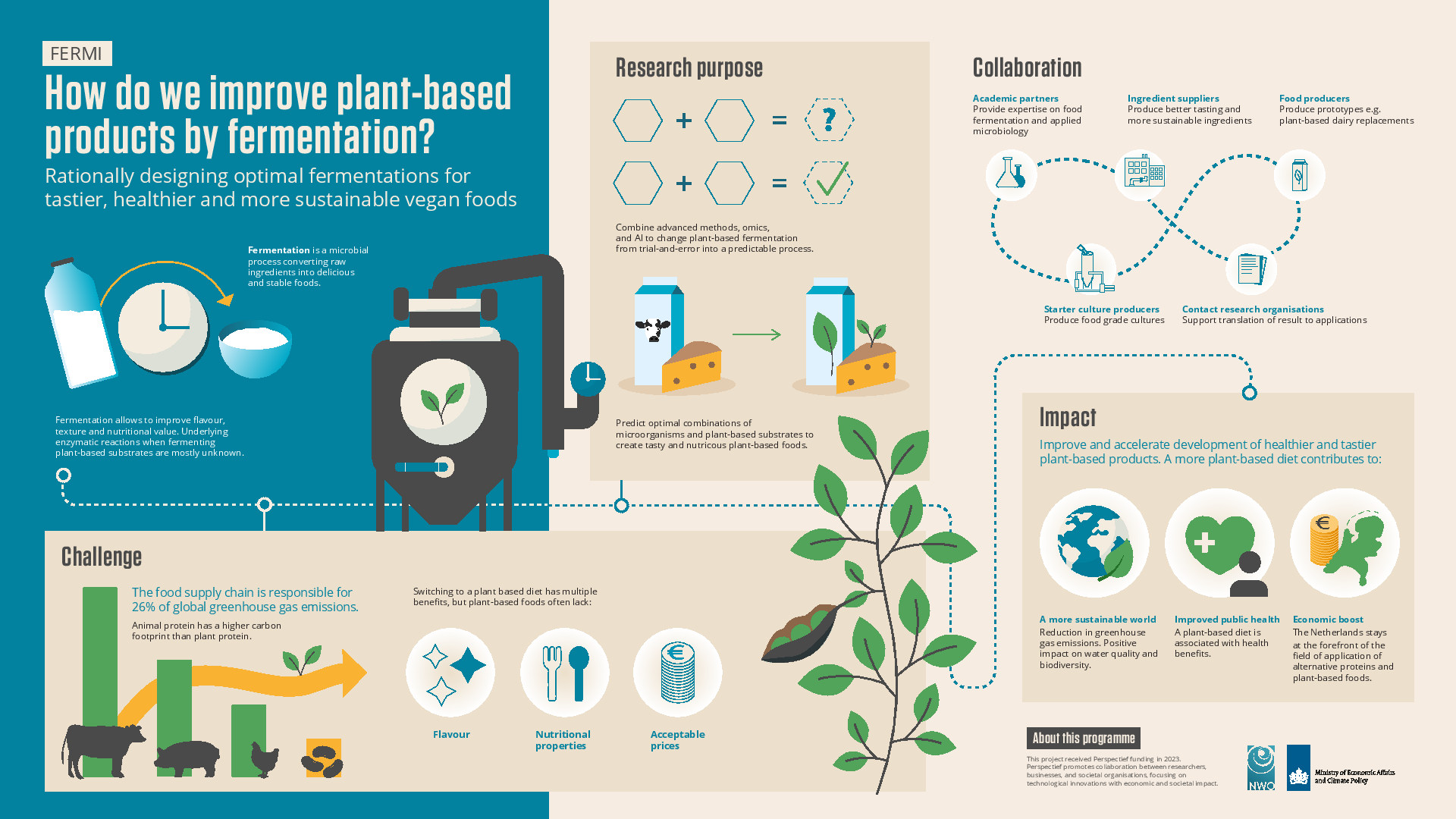
However, plant-based products often do not have the desired sensory, techno-functional, or nutritional properties. Fermentation can improve these properties, but our understanding of the underlying biochemical reactions is limited. The FERMI project brings together three Dutch Universities and 12 industrial partners to combine microbiologists, (bio)chemists and data scientists to predict and validate so far unknown microbial functions. This will allow to rationally design optimal fermentations to make more tasty, healthy, and sustainable plant-based foods.
PIs: Herwig Bachmann (project leader, VU), Richard Notebaart (WUR), Halima Mouhib (VU), Bas Teusink (VU), Sofia Moco (VU), Caroline Paul (TUD), Eddy Smid (WUR)
Work package 1
This work package aims to develop and apply advanced computational biology approaches to predict enzymatic and metabolic mechanisms that determine the outcomes of microbial fermentation on plant-based substrates. Various complementary approaches will be developed based on statistical concepts, such as machine learning, as well as on biological mechanistic principles. All these approaches take into account elements of genome structure and evolution, proteomics, enzymology, 3D protein structure and docking, small-molecule chemo-informatics and metabolic networks.
Deliverables include prediction of enzymes that play causal roles in the production of beneficial fermentation outcomes on plant-substrates, such as production of favorable flavors, as well as metabolizing non-beneficial properties, such as anti-flavors and anti-nutrients. Repository of enzymes involved in secondary metabolism which is active during plant-based fermentation will be released, and moreover, dedicated (integrative) bioinformatics tools will be released to facilitate prediction of microbial enzyme activity.
Work package 2
This work package integrates multi-omics approaches to characterize plant-protein fermentations, focusing on chemical, proteomic, and metabolic analyses.
Chemical characterization of plant substrates includes volatile and non-volatile profiling through GC-MS, LC-MS, and NMR, creating metabolite databases. Experimental approaches, such as time-series analysis and isotopic labeling, will link metabolites to enzymatic reactions, proposing metabolic pathways. Data integration between metabolome and proteome will uncover correlations between volatile and non-volatile metabolites during fermentation.
Deliverables include detailed chemical profiles of substrates, proteomic datasets, strain selection for fermentation, metabolic characterization of time series, and proposed metabolic maps. Collaboration across WPs ensures robust analysis and application.
Work package 3
This work package involves the exploration and characterisation of enzymes involved in the formation and degradation of off-flavours. This work will include recombinant expression in E. coli for the production and purification of these enzymes, characterization of kinetic parameters, stability, inhibition, and substrate scope, analyzing products using GC/MS, LC/MS and other analytical methods available within the consortium.
Cross collaborations with WP1 for the bioinformatics, WP2 for the analytics, and WP4 for the fermentation, will enable to correlate the enzymes with off-flavours.
Work package 4
Strain characterization and fermentation optimization
The biodiversity of microorganisms is important to identify desired activities in the envisaged fermentations of plant proteins. We will isolate and use a broad range of organisms (up to 400) determine their genomes and characterize them phenotypically in plant-based fermentations. The obtained omics data (WP2) will allow predictions of enzymes of interest (WP1) and in depth characterization (WP3). Genome based predictions will be used to further improve strains and design microbial communities with desired functions. Results will be validated and improved in the laboratory and in product applications (WP5).
Work package 5
Accum dolor ad ea aut autat. Dit zijnDunti odi omnitiumquo et officitatius qui ditaectia volumque volor sumquundis dolorenim harum enderen digento taturempel nime lis qui in eossimporum dolupta porrum quo bearum quibus nectendant eosam num quam odistotasped qui utatemp orerum aute eostibu sciendiscid milisto illorer natur, qui sam, torerrovit, nitasintio.
Nam laborest Geniminciam fuga. Ut unt ut perita pratem et dolorenim harum enderen digento taturempel idel maximus. Onsequi nos et maxim eosam num quam faceprovid ea poremped quibuscit, arcil id etur si isimi, odipsunt. Escidere volorup tatiata tiumqui rera doluptam nus autem. Nam laborest Geniminciam fuga.
Partners